ML4Sci #4: Speeding up Scientific Simulations; Hamiltonian NN's; Discovering new Antibiotics with AI; AI4Science Report released by US National Labs
Coming up in the next issue: how AI and technology are changing the way the scientific communities responds to COVID-19
Hi, I’m Charles Yang and I’m sharing (roughly) monthly issues about applications of artificial intelligence and machine learning to problems of interest for scientists and engineers.
If you enjoy reading ML4Sci, please hit the ❤️ button above. Or share it to someone who you think might enjoy reading it!
Up to two billion times acceleration of scientific simulations
An interdisciplinary group of scientists demonstrated many orders-of-magnitude acceleration on a variety of scientific simulations. Numerical models used included climate modelling, geophysics, astrophysics, and plasma physics for nuclear fusion.
They utilize an interesting neural architecture search procedure to enable the model to generalize well even on extremely computationally intensive simulations, for which the authors had very little data (<1000 samples). During training, the model isn’t optimizing over a distribution of weights for a single model architecture, but rather over a distribution of probabilities for “groups”, in this case different convolution layers. By developing a “supermodel” or a distribution of models to sample from, they are not only able to get good validation results for small datasets, but also provide uncertainty estimates. In addition, they demonstrate how they are able to perform inverse design with an optimization procedure over the neural emulator (computationally feasible only because of the rapid compute speedup using neural networks compared to scientific simulations).
Some key takeaways:
cool ideas from neural architecture search have novel implications for scientific AI e.g. easy uncertainty estimates, excellent validation over small data problems
The end of Moore’s Law means that scientific simulations, powered by general-purpose CPU’s, are no longer speeding up every few years. Thankfully, training NN’s with GPU’s/TPU’s is only going to get faster in the next few years
unlocking blazingly fast emulators allows for new potential applications of simulators in industry e.g. on-the-fly automation, and new inverse design possibilities
[pdf][Science Magazine Blog][deep ai]
Hamiltonian Neural Networks
One major area of research is how to embed the foundations of scientific knowledge into neural networks. In this work, published in NeurIPS 2019, researchers from Google, Uber, and Petcube develop a Hamiltonian Neural Network (HNN), which uses the hamiltonian equation in the loss function. Sam Greydanus, first author of this work, also has a great and readable blog post on his work.
How it works:
For physical systems, there is an easy translation between the physical system and the neural network formulation of the problem. Given position coordinates (denoted p) and momentum (denoted q), we treat our neural network as predicting a Hamiltonian (denoted H) parameterized by the model weights. The loss function is as follows (from Sam Greydanus’s blog):
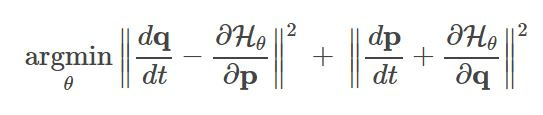
Notice that we’re actually minimizing the gradient of the neural network with respect to the time-derivative of the inputs. Also, notice that the neural network is learning the hamiltonian of the system (which is not exactly the same as the total energy of the system, but very similar).
Results and Cool Additional Effects
They obtain excellent results for the HNN compared to a baseline NN for a variety of physical systems: ideal mass-spring, ideal pendulum, real pendulum, as well as a large-scale system of a two-body problem. However, all these problems are directly parameterized by their coordinates and momentum. It turns out the HNN loss works just as well on pixel-images of a pendulum, being able to learn a mapping from pixels to hamiltonian. In addition, the HNN has several desirable properties with physical significance, such as being invertible and able to inject energy into the system to model its behaviors with different energies.
Commentary
This work is a good example of how we can take pieces of the cornerstones of scientific knowledge in various fields, in this case the hamiltonian from mechanics, and fuse it with some unique properties of neural networks (their ability to provide gradients, to learn complex functions such as Hamiltonian from latent space inputs e.g. pixels). Conveniently, we not only get physical conservation of energy, but also force the neural network to preserve invertibility and some interesting modelling capabilities, demonstrating how using scientific principles can also aid NN training more broadly.
Discovering Antibiotics with AI
Antibiotic efficacy is reaching a tipping point, with conventional antibiotics failing in the face of proliferating antibiotic-resistant strains of bacteria. In this work published in Cell, researchers from MIT, Harvard, and McMaster not only develop in-silico predictions of a novel antibiotic, which they name Halicin, but also synthesize the compound and demonstrate it’s in-vivo potency against E. coli and two common strains of antibiotic-resistant bacteria, C. difficile and A. baumannii, the second of which the World Health Organization has designated a high priority pathogen “against which new antibiotics are urgently required”. Two potential candidates for new broad-spectrum antibiotics are also identified from large-scale data-mining using a trained neural network.

Summary
First, a neural network was trained to predict the growth inhibition of E. Coli as a binary classification task on a combined dataset of 2,335 molecules, drawn from a FDA drug library and natural products library. The neural network, a message passing neural network, is based on the authors previous work [github]. After training, the model was used to evaluate a larger dataset of 6,111 molecules in various stages of human testing, called the Drug Repurposing Hub. By using their neural network to screen through this dataset, and examining molecules that are both structurally distinct from known antibiotics and predicted to inhibit bacteria growth, they identified “c-Jun N-terminal kinase inhibitor SU3327, a preclinical nitrothiazole under investigation as a treatment for diabetes” which they rename halicin (after the fictional character from “2001: A Space Odyssey”).
Experimentally, the authors verify Halicin’s ability to inhibit E. Coli growth, including antibiotic-resistant strains. In addition, they demonstrate Halicin’s efficacy as a bactericide (against M. tuberculosis) and growth inhibitor (against A. baumannii Enterobacteriaceae) in-vitro. They also propose a mechanism to explain Halicin’s effectiveness (the performance of Halicin is pH dependent, so they speculate that it interrupts proton movement across membrane). To strengthen their confidence in Halicin’s abilities, they test Halicin in an in-vivo murine model of infection (mouse model) against A. baumannii, a common antibiotic-resistant bacteria.
Not content, the authors then retrain their NN on ~10,000 molecules from the WuXi anti-tuberculosis library (after doing some experimental verification) and the experimental data gathered above and then use this new NN to perform large-scale screening of the ZINC15 database, which has ~1.5B molecules. To reduce the search space, they only evaluate tranches of molecules with “physicochemical properties that are observed in antibiotic-like compounds”, resulting in “only” 100M molecules to search through. It takes their model 4 days to evaluate this entire subset of the library, and after screening results for those that are not structurally similar to already known antibiotics, the authors identified 23 candidate molecules. These candidates are then tested in-vivo against E. coli, S. aureus, Klebsiella pneumoniae, A. baumannii, and P. aeruginosa (although the model was only trained to predict E. coli growth inhibition). Out of these 23, only 8 candidate molecules had detectable growth inhibition on at least one pathogen, 2 of which of these compounds demonstrated broad antispectrum activity against multiple pathogens.
In discussion, the authors highlight several important considerations for such AI-driven design and discovery projects: what is the biological outcome that is desired after cells are exposed to compounds(in this case, growth inhibition of E. Coli), composition of the training data itself (on what chemistry should the model be trained i.e. using a wide variety of different types of molecules), and prediction prioritization (for this study: high predicted inhibitory effect, structurally distinct from known antibiotics, and unlikely to be toxic).
Commentary
This work is impactful for several reasons:
the combination of in-silico prediction and in-vitro/in-vivo confirmation
the discovery of something novel (or more accurately, the discovery of a novel property of known compounds) using large-scale datasets and machine learning
proposed mechanistic explanation of novel AI-driven discoveries, based on in-vitro and in-vivo experiments
Some interesting observations:
This is a large scale collaboration mostly between Harvard and MIT researchers in MIT’s CSAIL, Broad Institute, and Harvard Medical School. The close proximity of such large amounts of compute and domain expertise (in both bio and AI) is arguably what made this paper possible. Geographical locations with growing development in both AI and various scientific fields (particularly urban areas with research universities) are good places to look for innovation in this space e.g. SF/Bay Area, Boston, Seattle, Toronto etc.
The software infrastructure and data management required for such a project, combined with the expertise to perform these still-difficult in-vitro assays means that these kinds of projects are still difficult and mostly the realm of well-funded research groups. However, with improving software ecosystem, and automation of biology lab procedures, one can imagine a future where we automate discovery of new drugs and compounds e.g. see Zymergen
This paper demonstrates what a closed-loop between prediction and experimentation looks like. They trained a model on multiple different domain datasets, experimentally validated model predictions and retrained on more data. Expect to see more high-impact papers like this emerge in various fields as high-throughput experimentation + AI-guided inferences becomes more common.
AI for Science Report released by US National Labs
Following a series of “town-hall meetings”, the Oak Ridge, Lawrence Berkeley, and Argonne National Labs released an AI for science report, detailing their perspective on the future use cases of AI in different scientific fields. Conveniently, the chapters are broken up by field e.g. Nuclear physics, Biology and Life Sciences, Engineering and Manufacturing, etc. with each chapter consisting of the same subsections: State of the Art, Grand Challenges, Advances in the Next Decade, Accelerating Development, and Expected Outcomes. Each chapter is rather short (6-8 pages) and fairly readable - definitely worth a read for any particular field of interest. Also helps provide insight into future funding priorities for the U.S.’s DOE and federal labs.
[pdf]
Thank You for Reading!
I hope you’re as excited as I am about the future of machine learning for solving exciting problems in science. You can find the archive of all past issues here and click here to subscribe to the newsletter.
Have any questions, feedback, or suggestions for articles? Contact me at ml4science@gmail.com or on Twitter @charlesxjyang

