ML4Sci #28: DeepMind "solves" protein folding
In the News: AI for Archaeology and Deep Learning as Black Magic
Hi, I’m Charles Yang and I’m sharing (roughly) weekly issues about applications of artificial intelligence and machine learning to problems of interest for scientists and engineers.
If you enjoy reading ML4Sci, send us a ❤️. Or forward it to someone who you think might enjoy it!
As COVID-19 continues to spread, let’s all do our part to help protect those who are most vulnerable to this epidemic. Wash your hands frequently (maybe after reading this?), wear a mask, check in on someone (potentially virtually), and continue to practice social distancing.
First, some housekeeping. The team I work with at Lawrence Berkeley National Lab (LBL) has finally published our paper on “Interpretable Forward and Inverse Design of Particle Spectral Emissivity” in Cell Reports Physical Science as an open-access article (of course, the rough draft has been on arxiv for 10 months now). In short, we use a ML paper from 1998, adapt it to the unique data constraints of (optical) numerical simulations, to achieve interpretable inverse design with decision trees. This work was also part of our initial proposal to the ARPA-E DIFFERENTIATE program, for which we are now in round 2 of funding. I enjoyed this work because I think it highlights a lot of the themes we talked about in the previous edition of ML4Sci: different priorities and constraints found in scientific problems lead to retooled methods and novel use-cases, particularly as we focused on interpretability and explainability.
I’m still experimenting with the format of this newsletter (though one would think I’d have figured something out by now 🙄). Since there’s been a lot of movement in the past couple weeks, I’m going to try a lighter, more broad style of coverage, rather than narrowing in on a few papers. I think I’ll stick to this format more often, while doing more in-depth coverage of a few (really good) papers every once in a while. Let me know what you think! (I also came up with snazzier subsection headings)
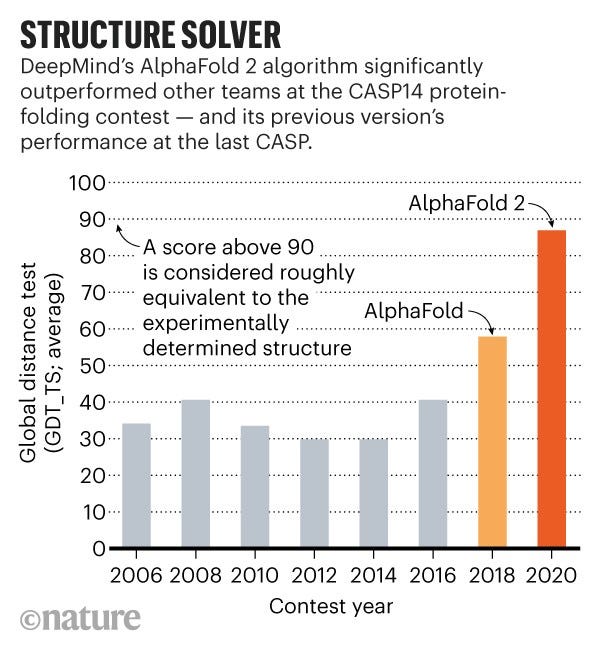
The biggest news this week is that Google’s DeepMind has “solved” protein folding this past week. Ok, not exactly.

A bit of a recap: DeepMind submitted their model AlphaFold last year to the annual Critical Assessment of Structure Prediction (CASP) competition, an annual challenge to predict protein structures, and it blew away the field, taking first place by a clear margin and beating out established computational biologist groups. This year, DeepMind submitted an improved version of the model that essentially “solved the competition” (see figure below from NatureNews)
Now, I don’t want to be a nay-sayer; this work is very impressive and I don’t think it’s too overboard to say “protein folding has its ‘ImageNet moment’”. People used to think it would be impossible for models to have human-level accuracy at recognizing images; then they thought it would be impossible for models to predict protein structures. Both of these have clearly been demonstrated now in short order.
But, as Vishal Gulati writes, “compared to the problem of protein folding, CASP is a game. It is a very hard game but it is a reduced problem set which helps us train our tools and standardize performance”. Similarly, after the 2012 “ImageNet moment”, we found out computer vision models suffer from adversarial attacks and poor generalizability in the real world. Derek Lowe also provides a clear-eyed assessment of the implications of DeepMind’s AlphaFold 2 for the clinical treatment pipeline in his Science Magazine blog (tldr; the human body is complicated and things regularly fail in clinical trial for unknown reasons).
So in short, an impressive technical demonstration by DeepMind on a difficult problem that nobody thought would be solved in such a short period of time. But the difficult work of science remains, albeit with a new tool in the toolkit. Of course, scarcely anyone, in between hyperventilated breathing about how AI has solved a broad swath of biology, is wondering about what the implications of the fact that a large tech company is leveraging AI expertise to begin making advances in what could be used for basic clinical research…
Department of Machine Learning
Kaggle(acquired by Google) releases its annual survey of data scientists. Survey respondents are disproportionately men, self-learners with college degrees, and from the US/India. They tend to use AWS, linear regression (!), and scikit-learn.
🔮“Black Magic in Deep Learning”: turns out hyperparameter tuning ability in deep learning is still a function of expertise, based on experiments with real researchers. It seems deep learning is still more of an art than a science.
A twitter thread on deep learning training tricks that you can only find in appendices:



Also on Twitter this week: Google forces out AI ethics researcher Timnit Gebru over a drafted publication criticizing large NLP models used by tech comapnies. At the very least, this is an example of the distortion in academic research, in this case AI ethics, that occurs when the bulk of talented researchers are at private companies
Near-Future Science
Semi-supervised learning to accelerate materials design, written by ML4Sci subscriber and PhD student @UPenn, Nathan Frey
🤖🧪IBM’s RoboRXN is an AI-powered “remotely accessible, fully autonomous” chemistry platform [website][youtube][github]
💻From Nature Review Physics: “The Emerging Commercial Landscape of Quantum Computing”
[NYT] Nice coverage of some of AI-Science, especially around the new NSF centers for AI and Physics
⚒️🪨[NYT] Archaeologists are using convolutional neural networks (and nobody is losing their jobs). AI will change the way we do science, but it is not yet replacing anything/anyone
🎈Google’s Loon Project published a 🔒paper in Nature[NatureNews] on using reinforcement learning to guide stratospheric balloons, which can be used for environmental monitoring or surveillance. Nicely captures some of the themes we talked about in the last newsletter: AI is affecting every aspect of science in unexpected ways, like enabling us to collect data more efficiently, and the dominance of tech companies in this space.
The Science of Science
Springer Nature reveals open-access plan - it will cost authors $11,000 to publish open-access. Of course, they could always publish on Arxiv for free and submit to ICLR for essentially nothing, but unfortunately, academia today doesn’t reward work published on Arxiv (Arxiv is also practically bankrupt). Open-access is a hard problem that will require institutional change and this news is….a step toward…. something.
eLife has implemented a “Publish, then Review” model. Quite an incredible step forward, but it still leaves unanswered the question of how preprint servers will pay for themselves. From the abstract:
From July 2021 eLife will only review manuscripts already published as preprints, and will focus its editorial process on producing public reviews to be posted alongside the preprints.
[TheGradient] How can we improve Peer Review in NLP
A nice review of recent peer review experiments run by ML conferences and some nice quotes
“peer review is fundamentally an annotation task”
Reviewers use heuristics like non-native english(❌), no deep learning(❌), no state-of-the-art(❌), etc.
“NeurIPS 2014 conducted an experiment where 10% of submissions were reviewed by two independent sets of reviewers. These reviewers disagreed on 57% of accepted papers (Price, 2014). In other words, if you simply re-ran the reviewer assignment with the same reviewer pool, over half of accepted NeurIPS papers would have been rejected!” (emphasis their own)
It is difficult to publish meta-reviews of peer review because nobody is incentivized to publish and do such research. As the authors put it, “NLP peer review… prevents research on NLP peer review”
🌎Out in the World of Tech
Open letter from content moderators at Facebook regarding working conditions during the pandemic
🤦♀️Gender Bias in Image Recognition Systems. From the abstract:
In this article, we evaluate potential gender biases of commercial image recognition platforms using photographs of U.S. members of Congress and a large number of Twitter images posted by these politicians. Our crowdsourced validation shows that commercial image recognition systems can produce labels that are correct and biased at the same time as they selectively report a subset of many possible true labels. We find that images of women received three times more annotations related to physical appearance. Moreover, women in images are recognized at substantially lower rates in comparison with men. We discuss how encoded biases such as these affect the visibility of women, reinforce harmful gender stereotypes, and limit the validity of the insights that can be gathered from such data
[NatureNews] “Ethical questions that haunt facial recognition researchers”, along with a nice survey of facial recognition researchers. Maybe they should thinking about working in ML4Sci!
Policy and Regulation
[Nat’l Bureau of Economics Research] “How to Talk When a Machine is Listening” Algorithmic scraping of corporate disclosures for sentiment analysis is changing the language corporations use in these reports. First we trained the machines, then the machines trained us.
Thanks for Reading!
I hope you’re as excited as I am about the future of machine learning for solving exciting problems in science. You can find the archive of all past issues here and click here to subscribe to the newsletter.
Have any questions, feedback, or suggestions for articles? Contact me at ml4science@gmail.com or on Twitter @charlesxjyang


Thanks for the links to more sober analyses of AlphaFold, Google's PR dept really went overboard on this one.